Tutorial¶
This simple guide can help you start working with Tree-AMP.
See also
- User guide
For an in-depth explanation of the package
- Examples
For a gallery of standalone examples
- arXiv preprint
For the research article
Building your first model¶
Creating a variable¶
Let us start by creating our first variable tramp.variables.SISOVariable V of size N drawn from a priors.GaussBernoulliPrior separable prior with sparsity rho
>>> from tramp.variables import SISOVariable as V
>>> from tramp.priors import GaussBernoulliPrior
>>> N, rho = 100, 0.1
>>> prior = GaussBernoulliPrior(size=N, rho=rho)
>>> var_x = prior @ V("x")
The opeartor @ allows to assign a variable x drawn from a prior distribution, and in general to concatenate modules together.
Draw a matrix from an ensemble¶
Creating a matrix W instance of the tramp.ensembles.GaussianEnsemble of size M x N can be done by
>>> from tramp.ensembles import GaussianEnsemble
>>> M, N = 200, 100
>>> ensemble = GaussianEnsemble(M, N)
>>> W = ensemble.generate()
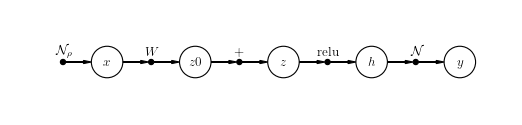
Constructing a linear channel¶
The variable x can be mutliplied by the matrix W using the channels.LinearChannel.
The result can be casted in a new variable z0 = W x with W ~ ensemble and x ~ prior.
>>> from tramp.channels import LinearChannel
>>> model = prior @ V("x") @ LinearChannel(W=W, name='W') @ V("z0")
Adding other modules¶
As many module as desired can be added to the previous model. Each module must be followed by a variable V.
For example a tramp.channels.BiasChannel with value b with intermediate variabel z,
followed by a tramp.channels.ReluChannel outputing a variable h.
>>> from tramp.channels import BiasChannel, ReluChannel
>>> b = 0.1
>>> model = model @ BiasChannel(bias=b) @ V("z")
>>> model = model @ ReluChannel() @ V("h")
Adding observations¶
Observations tramp.SILeafVariable O can be added to the the model.
For example model may go through a noisy tramp.channels.GaussianChannel with variance var and outputs a variable y
>>> from tramp.variables import SILeafVariable as O
>>> from tramp.channels import GaussianChannel
>>> var = 1e-2
>>> model = model @ GaussianChannel(var=var) @ O("y")
EP/SE in the Bayes-optimal scenario¶
Let us illustrate how to run EP or SE in the Bayes-optimal case and on a more complex model: a sparse gradient prior.
Build the model¶
Creating a variable x of size N sampled from a tramp.priors.GaussianPrior with n_next=2 successors can be done with a tramp.variables.SIMOVariable V
>>> from tramp.variables import SIMOVariable as V
>>> from tramp.priors import GaussianPrior
>>> N = 100
>>> prior_x = GaussianPrior(size=N, var=1) @ V(id="x", n_next=2)
which can be connected to a tramp.channels.GaussianChannel with y observations and a sparse gradient constraint on x.
This constraint can be built with a new variable x' connected to n_prev=2 predecesors: a tramp.channels.GradientChannel connected to x and a tramp.priors.GaussBernoulliPrior with sparsity rho.
>>> from tramp.channels import GradientChannel, GaussianChannel
>>> from tramp.priors import GaussBernoulliPrior
>>> from tramp.variables import SISOVariable as V, MILeafVariable
>>> rho = 0.04
>>> var = 0.1
>>> x_shape = (N,)
>>> grad_shape = (1,) + x_shape
>>> # Create the gaussian channel and the observations y
>>> channel_y = GaussianChannel(var=var) @ O("y")
>>> # Create the sparse gradient constraint and the new variable x'
>>> channel_grad = (GradientChannel(shape=x_shape) + GaussBernoulliPrior(size=grad_shape, rho=rho)) @ MILeafVariable(id="x'", n_prev=2)
>>> # Plug the two channels to the variables x
>>> model = prior_x @ ( channel_y + channel_grad )
>>> # Build the model
>>> model = model.to_model()
>>> # Show the model
>>> model.plot()
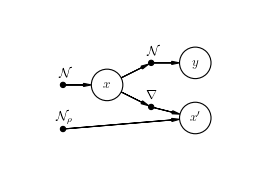
Teacher-Student scenario¶
In a tramp.experiments.BayesOptimalScenario, ground truth samples of all the variables contained in the model are drawn (namely x, x', y ).
The scenario tries then to infer the variables x_ids from the observations y and knowing the structure and the parameters of the model (rho, var).
The scenario is setup with a .setup() method where an optional seed can be used for reproducibility.
>>> from tramp.experiments import BayesOptimalScenario
>>> x_ids = ["x", "x'"]
>>> scenario = BayesOptimalScenario(model, x_ids=x_ids)
>>> scenario.setup(seed=42)
Run Expectation Propagation (EP)¶
EP can be run on the above scenario with the method .run_ep().
Optional maximum number of iterations max_iter and damping on all variables to help convergence can be added.
Different callback can be used such as tramp.algos.EarlyStoppingEP if precision tol is reached.
>>> from tramp.algos import EarlyStoppingEP
>>> scenario.run_ep(max_iter=1000, damping= 0.1, callback=EarlyStoppingEP(tol=1e-2))
To use multiple callbacks at a time, you need to use tramp.algos.JoinCallback:
>>> from tramp.algos import JoinCallback, EarlyStoppingEP, TrackOverlaps, TrackEstimate
>>> track_overlap = TrackOverlaps(true_values=scenario.x_true)
>>> track_estimate = TrackEstimate()
>>> callback = JoinCallback([track_overlap, track_estimate, EarlyStoppingEP(tol=1e-12)])
Run State Evolution (SE)¶
SE can be performed simply with the run_se() method
>>> scenario.run_se()
Todo
Continue the tutorial