Note
Click here to download the full example code
Raccoon denoising¶
from tramp.algos import ConstantInit
from tramp.priors.base_prior import Prior
from scipy.misc import face
from scipy.stats import laplace
from tramp.experiments import TeacherStudentScenario
from tramp.priors import BinaryPrior, GaussianPrior, GaussBernoulliPrior, MAP_L21NormPrior
from tramp.channels import Blur2DChannel, GaussianChannel, GradientChannel
from tramp.variables import SIMOVariable, MILeafVariable, SISOVariable as V, SILeafVariable as O
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
plt.rc("text", usetex=True)
plt.rc('font', family='serif', size=14)
Plotting functions
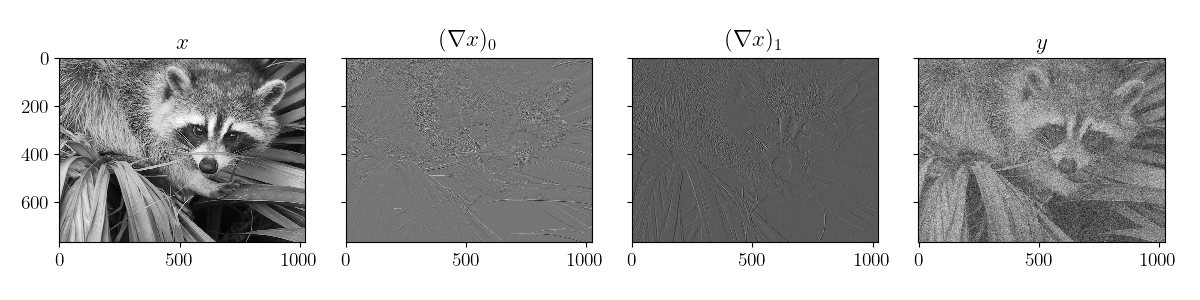
def plot_data(x_data, y=None):
n_axs = 3 if y is None else 4
fig, axs = plt.subplots(1, n_axs, figsize=(3*n_axs, 3), sharey=True)
axs[0].imshow(x_data["x"], cmap="gray")
axs[0].set(title=r"$x$")
axs[1].imshow(x_data["x'"][0], cmap="gray")
axs[1].set(title=r"$(\nabla x)_0$")
axs[2].imshow(x_data["x'"][1], cmap="gray")
axs[2].set(title=r"$(\nabla x)_1$")
if y is not None:
axs[3].imshow(y, cmap="gray")
axs[3].set(title=r"$y$")
fig.tight_layout()
def compare_hcut(x_true, x_pred, y=None, h=25):
fig, axs = plt.subplots(1, 2, figsize=(9, 3), sharex=True, sharey=False)
axs[0].plot(x_true["x"][:, h], label="true")
axs[0].plot(x_pred["x"][:, h], label="pred")
if y is not None:
axs[0].plot(y[:, h], ".", color="gray", label="y")
axs[0].legend()
axs[0].set(title=r"$x$")
axs[1].plot(x_true["x'"][0, :, h], label="true")
axs[1].plot(x_pred["x'"][0, :, h], label="pred")
axs[1].legend()
axs[1].set(title=r"$(\nabla x)_0$")
fig.tight_layout()
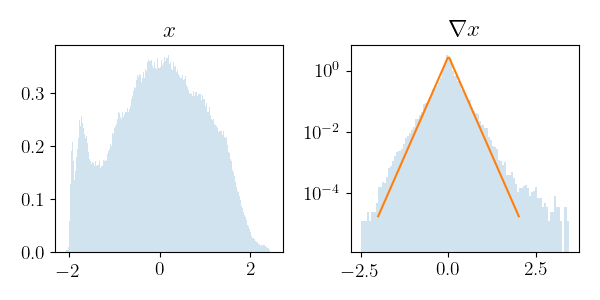
def plot_histograms(x_data):
fig, axs = plt.subplots(1, 2, figsize=(6, 3), sharey=False)
axs[0].hist(
x_data["x"].ravel(), bins=251,
density=True, histtype='stepfilled', alpha=0.2
)
axs[0].set(title=r"$x$")
# hyperparam for grad x
x_grad = x_data["x'"].ravel()
nonzero = np.abs(x_grad) > 1e-3
grad_rho = nonzero.mean()
grad_var = x_grad[nonzero].var()
grad_loc, grad_gamma = laplace.fit(x_grad[nonzero])
print(
f"grad_rho={grad_rho:.3f} grad_var={grad_var:.3f} grad_gamma={grad_gamma:.3f}")
# compare laplace fit and empirical distribution
fitted = laplace(loc=0, scale=grad_gamma)
axs[1].hist(
x_grad[nonzero], bins=100, log="y",
density=True, histtype='stepfilled', alpha=0.2
)
t = np.linspace(-2, 2, 100)
axs[1].plot(t, fitted.pdf(t))
axs[1].set(title=r"$\nabla x$")
fig.tight_layout()
Build the teacher
class RaccoonPrior(Prior):
def __init__(self):
x = face(gray=True).astype(np.float32)
x = (x - x.mean())/x.std()
self.x = x
self.size = x.shape
def sample(self):
return self.x
prior = RaccoonPrior()
x_shape = prior.size
noise = GaussianChannel(var=0.5)
grad_shape = (2,) + x_shape
teacher = (
prior @ SIMOVariable("x", n_next=2) @ (
GradientChannel(x_shape) @ O("x'") + noise @ O("y")
)
).to_model()
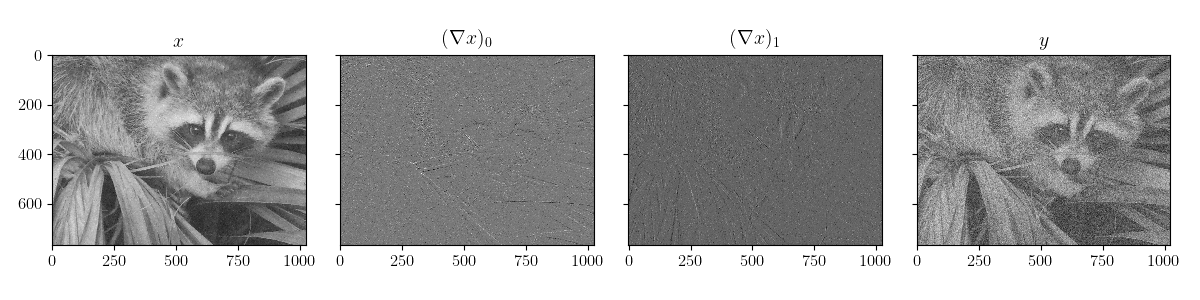
sample = teacher.sample()
plot_histograms(sample)
plot_data(sample, sample["y"])
Out:
grad_rho=0.896 grad_var=0.065 grad_gamma=0.166
Sparse gradient denoising
grad_shape = (2,) + x_shape
sparse_grad = (
GaussianPrior(size=x_shape) @
SIMOVariable(id="x", n_next=2) @ (
noise @ O("y") + (
GradientChannel(shape=x_shape) +
GaussBernoulliPrior(size=grad_shape, var=0.7, rho=0.9)
) @
MILeafVariable(id="x'", n_prev=2)
)
).to_model()
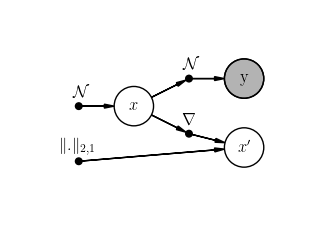
scenario = TeacherStudentScenario(teacher, sparse_grad, x_ids=["x", "x'"])
scenario.setup(seed=1)
scenario.student.plot()
_ = scenario.run_ep(max_iter=100, damping=0.1)
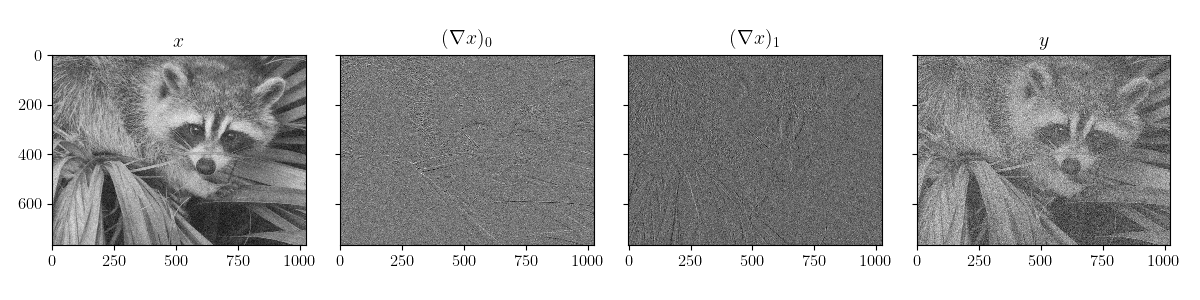
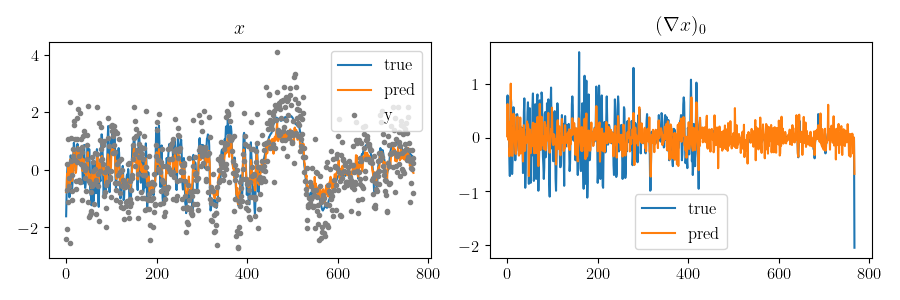
plot_data(scenario.x_pred, scenario.observations["y"])
compare_hcut(scenario.x_true, scenario.x_pred,
scenario.observations["y"], h=20)
Total variation denoising We need to set initial conditions a = b = 1. For a = b = 0 ExpectationPropagation diverges.
tv_denoiser = (
GaussianPrior(size=x_shape) @
SIMOVariable(id="x", n_next=2) @ (
noise @ O("y") + (
GradientChannel(shape=x_shape) +
MAP_L21NormPrior(size=grad_shape, gamma=1)
) @
MILeafVariable(id="x'", n_prev=2)
)
).to_model()
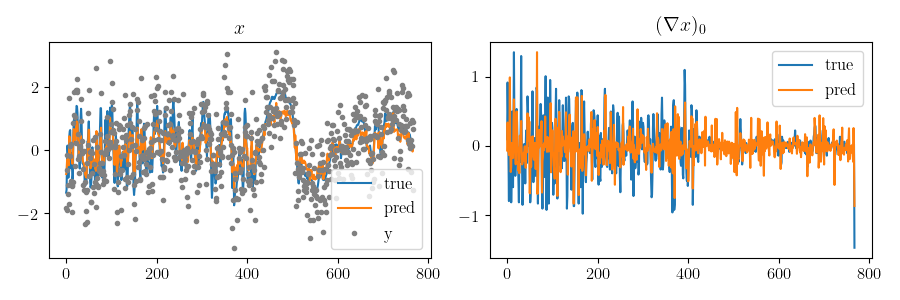
scenario2 = TeacherStudentScenario(teacher, tv_denoiser, x_ids=["x", "x'"])
scenario2.setup(seed=1)
scenario2.student.plot()
_ = scenario2.run_ep(max_iter=100, damping=0,
initializer=ConstantInit(a=1, b=1))
plot_data(scenario2.x_pred, scenario2.observations["y"])
compare_hcut(scenario2.x_true, scenario2.x_pred,
scenario2.observations["y"], h=25)
Out:
/Users/antoinebaker/Documents/projects/ml_physics/tramp/tramp/priors/map_L21_norm_prior.py:53: UserWarning: MAP_L21NormPrior.sample not implemented return zero array as a placeholder
"MAP_L21NormPrior.sample not implemented "
Total running time of the script: ( 0 minutes 49.381 seconds)